KatGs are enzymes found in
many bacteria as well as several fungi and protists. As the name
suggests, these enzymes have the ability to decompose hydrogen
peroxide by two primary mechanisms: catalase and
peroxidase. A central component of the defensive response of
plants and animals to invading
pathogens is the production of copious amounts of hydrogen
peroxide. Consequently, KatG figures prominently in the
antioxidant defenses of several notorious pathogens. Examples
include E. coli O157:H7 (a highly virulent food-borne
pathogen), Yersina pestis (the cause of bubonic plague),
and Magnaporthe grisea (the cause of rice blast
diesease and a major threat to world food security).
Interestingly, KatG is the sole catalase-active enzyme produced
by M. tuberculosis, an organism that survives and
propagates in the phagolysosomes of neutrophils and macrophages.
In addition, M. tuberculosis KatG is the enzyme
responsible for activating the antitubercular pro-drug
isoniazid, one of the front-line agents used to fight
tuberculosis. Consequently, mutations affecting the katG
gene are a prominent underlying cause
for resistance of numerous strains of Tb to isoniazid
chemotherapy. Clearly, there are several biomedical benefits to
be derived from understanding the connection between KatG
structure and function.
The bifunctional capability
of catalase-peroxidases is an anomaly. Typical catalases are not
especially robust peroxidases, and canonical peroxidases (e.g.,
cytochrome c peroxidase) are abysmal as catalases.
Interestingly, KatG is a member of the peroxidase-catalase
superfamily along with enzymes like cytochrome c
peroxidase. This is immediately obvious when one compares
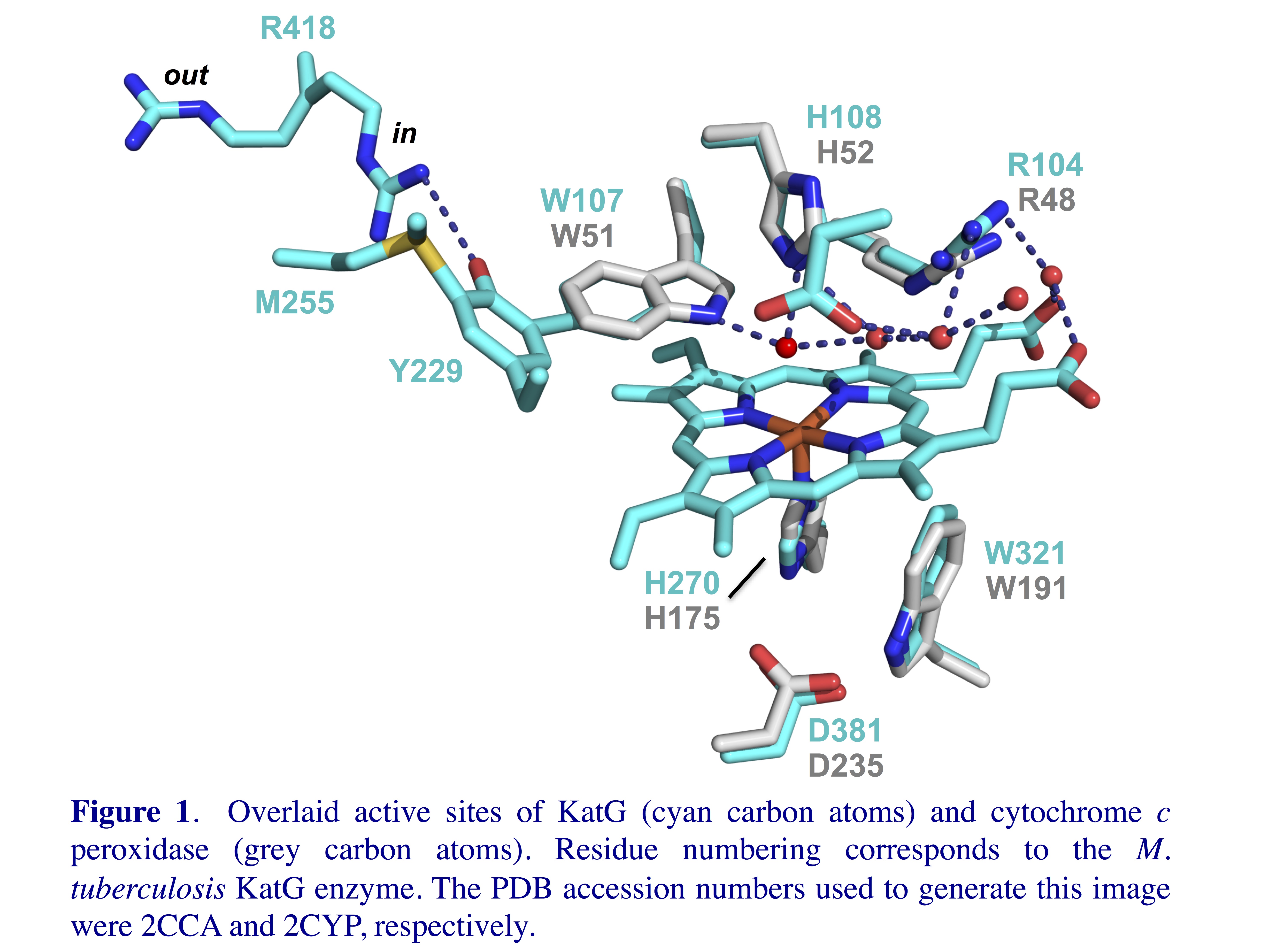
the active sites of these enzymes (Figure 1). The active
sites of KatG and cytochrome c peroxidase, one of its
closest relatives, are superimposable. Even the much more
distantly related manganese peroxidase enzyme has essentially
the same active site features with only two active site
tryptophan residues replaced by phenylalanines. Despite the great similarity of their
active sites, KatG is the only member of this superfamily to
show appreciable catalase activity.
What gives KatG the ability to carry out such robust catalase activity compared to its peroxidase relatives? All KatGs examined to date show a novel methionine-tyrosine-tryptophan (MYW) covalent adduct. With substitutions to any of the members of the adduct, KatG loses all catalase activity but still shows comparable if not enhanced peroxidase activity. These data suggest the MYW adduct is a cofactor for KatGs catalase activity. Together, the distinct peroxidase-like active site (as opposed to that of a typical catalase) and the presence of a novel cofactor suggest that KatG operates by novel catalase mechanism. Indeed, a radical centered on the MYW cofactor, a perhydroxy derivative of the MYW tryptophan, and a ferri-superoxo heme center have all been put forward as potential intermediates for this novel mechanism. In addition, one must ask how KatG manages the interplay of its two major catalytic activities, catalase and peroxidase. From its discovery, it has been presumed that the two are mutually antagonistic, but we have observed that peroxidatic electron donors actually stimulate KatG catalase activity by several fold. Our data point to a synergistic cooperation between the two activities that emerges because of two pathways for intramolecular electron transfer. What is most striking about this unexpected synergistic cooperation between catalase and peroxidase acitivities, is that it expands the capacity and range of KatG to respond to threats from hydrogen peroxide. This is particularly important in the context of host innate immune responses.